peroxide/numerical/optimize.rs
1//! To optimize parametric model (non-linear regression)
2//!
3//! ## `Optimizer` structure
4//!
5//! ### Declaration
6//!
7//! ```rust
8//! extern crate peroxide;
9//! use peroxide::fuga::*;
10//! use std::collections::HashMap;
11//!
12//! pub struct Optimizer<F>
13//! where F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>> {
14//! domain: Vec<f64>,
15//! observed: Vec<f64>,
16//! func: Box<F>,
17//! param: Vec<AD>,
18//! max_iter: usize,
19//! error: f64,
20//! method: OptMethod,
21//! option: HashMap<OptOption, bool>,
22//! hyperparams: HashMap<String, f64>,
23//! }
24//! ```
25//!
26//! ### Method (Should fill)
27//!
28//! * `new` : Declare new Optimizer. **Should be mutable**
29//! * `set_init_param` : Input initial parameter
30//! * `set_max_iter` : Set maximum number of iterations
31//! * `set_method` : Set method to optimization
32//!
33//! ### Method (Optional)
34//!
35//! * `get_domain` : Get domain
36//! * `get_error` : Root mean square error
37//! * `get_hyperparam` : Get hyperparameter
38//! * `set_lr` : Set learning rate (For `GradientDescent`)
39//! * `set_lambda_init` : Set initial value of lambda (For `LevenbergMarquardt`)
40//! * `set_lambda_max` : Set maximum value of lambda (For `LevenbergMarquardt`)
41//!
42//! ### Method (Generate result)
43//!
44//! * `optimize` : Optimize
45//!
46//! ## Example
47//!
48//! * Optimize $y = x^n$ model with $y = x^2$ with gaussian noise.
49//!
50//! ```rust
51//! #[macro_use]
52//! extern crate peroxide;
53//! use peroxide::fuga::*;
54//!
55//! fn main() {
56//! // To prepare noise
57//! let normal = Normal(0f64, 0.1f64);
58//! let normal2 = Normal(0f64, 100f64);
59//!
60//! // Noise to domain
61//! let mut x = seq(0., 99., 1f64);
62//! x = zip_with(|a, b| (a + b).abs(), &x, &normal.sample(x.len()));
63//!
64//! // Noise to image
65//! let mut y = x.fmap(|t| t.powi(2));
66//! y = zip_with(|a, b| a + b, &y, &normal2.sample(y.len()));
67//!
68//! // Initial parameter
69//! let n_init = vec![1f64];
70//! let data = hstack!(x.clone(), y.clone());
71//!
72//! // Optimizer setting
73//! let mut opt = Optimizer::new(data, quad);
74//! let p = opt.set_init_param(n_init)
75//! .set_max_iter(50)
76//! .set_method(LevenbergMarquardt)
77//! .set_lambda_init(1e-3) // Optional: Set initial value of lambda (Only for `LevenbergMarquardt`)
78//! .set_lambda_max(1e+3) // Optional: Set maximum bound of lambda (Only for `LevenbergMarquardt`)
79//! .optimize();
80//! p.print(); // Optimized parameter
81//! opt.get_error().print(); // Optimized RMSE
82//!
83//! // Plot
84//! //#[cfg(feature = "plot")]
85//! //{
86//! // let z = quad(&x, p.to_ad_vec()).unwrap().to_f64_vec();
87//! // let mut plt = Plot2D::new();
88//! // plt.set_domain(x)
89//! // .insert_image(y) // plot data
90//! // .insert_image(z) // plot fit
91//! // .set_legend(vec!["Data", "Fit"])
92//! // .set_title("Test ($y=x^2$ with noise)")
93//! // .set_path("example_data/lm_test.png")
94//! // .set_marker(vec![Point, Line])
95//! // .savefig().expect("Can't draw a plot");
96//! //}
97//! }
98//!
99//! fn quad(x: &Vec<f64>, n: Vec<AD>) -> Option<Vec<AD>> {
100//! Some(
101//! x.clone().into_iter()
102//! .map(|t| AD1(t, 0f64))
103//! .map(|t| t.pow(n[0]))
104//! .collect()
105//! )
106//! }
107//! ```
108//!
109//! 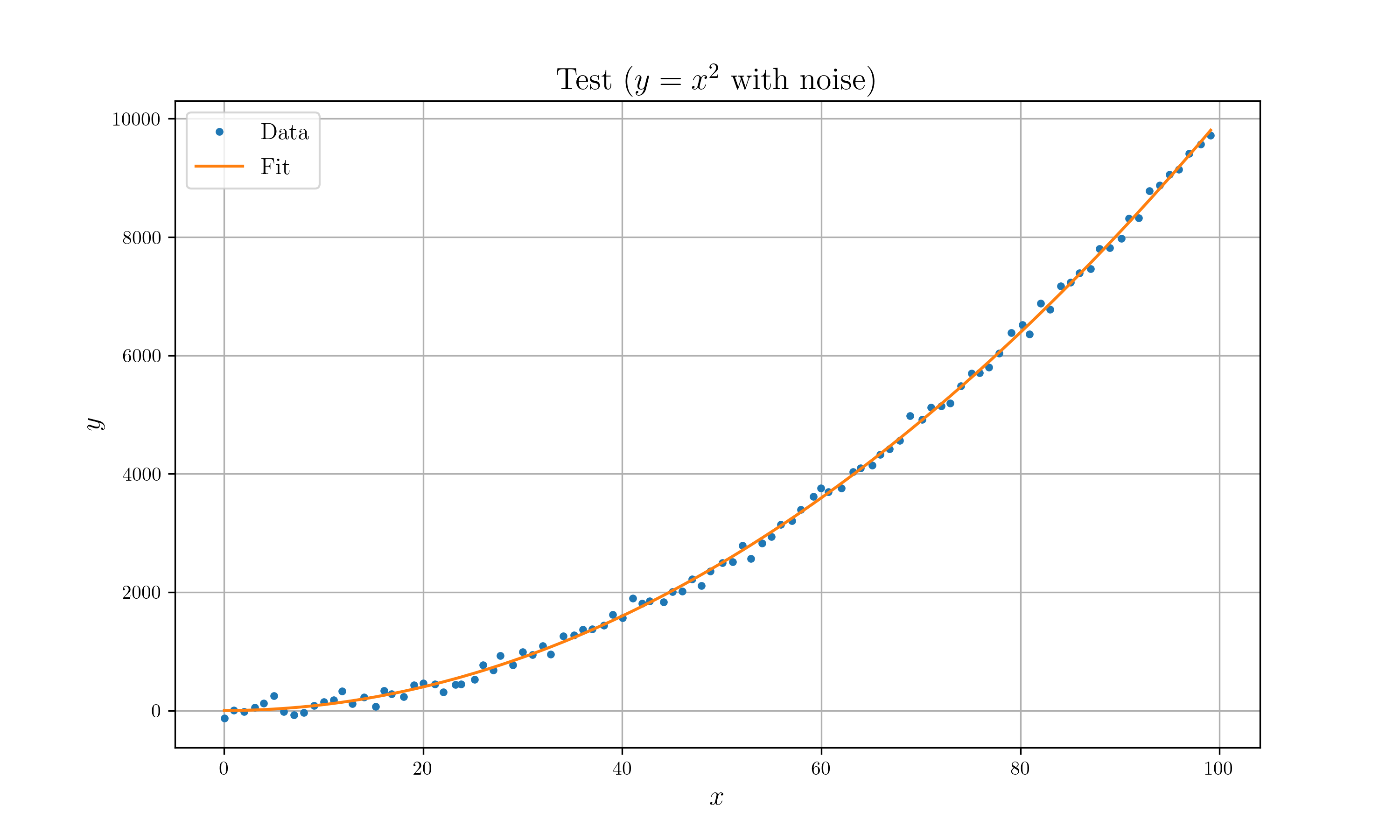
110
111pub use self::OptMethod::{GaussNewton, GradientDescent, LevenbergMarquardt};
112use self::OptOption::{InitParam, MaxIter};
113use crate::fuga::ConvToMat;
114use crate::numerical::utils::jacobian;
115use crate::structure::ad::{ADVec, AD};
116use crate::structure::matrix::Matrix;
117use crate::traits::matrix::{LinearAlgebra, MatrixTrait};
118use crate::util::useful::max;
119use std::collections::HashMap;
120
121#[derive(Debug, Clone, Copy)]
122pub enum OptMethod {
123 GradientDescent,
124 GaussNewton,
125 LevenbergMarquardt,
126}
127
128#[derive(Debug, Clone, Copy, PartialOrd, PartialEq, Eq, Hash)]
129pub enum OptOption {
130 InitParam,
131 MaxIter,
132}
133
134/// Optimizer for optimization (non-linear regression)
135///
136/// # Methods
137/// * Gradient Descent
138/// * Gauss Newton
139/// * Levenberg Marquardt
140///
141/// # Caution
142/// * `func` should be boxed. (This allows more generic function)
143pub struct Optimizer<F>
144where
145 F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>>,
146{
147 domain: Vec<f64>,
148 observed: Vec<f64>,
149 func: Box<F>,
150 param: Vec<AD>,
151 max_iter: usize,
152 error: f64,
153 method: OptMethod,
154 option: HashMap<OptOption, bool>,
155 hyperparams: HashMap<String, f64>,
156}
157
158impl<F> Optimizer<F>
159where
160 F: Fn(&Vec<f64>, Vec<AD>) -> Option<Vec<AD>>,
161{
162 pub fn new(data: Matrix, func: F) -> Self {
163 let mut default_option: HashMap<OptOption, bool> = HashMap::new();
164 default_option.insert(InitParam, false);
165 default_option.insert(MaxIter, false);
166
167 Optimizer {
168 domain: data.col(0),
169 observed: data.col(1),
170 func: Box::new(func),
171 param: vec![],
172 max_iter: 0,
173 error: 0f64,
174 method: LevenbergMarquardt,
175 option: default_option,
176 hyperparams: HashMap::new(),
177 }
178 }
179
180 /// Get domain
181 pub fn get_domain(&self) -> Vec<f64> {
182 self.domain.clone()
183 }
184
185 /// Get error
186 pub fn get_error(&self) -> f64 {
187 self.error
188 }
189
190 /// Get hyperparameter (learning rate or lambda or etc.)
191 pub fn get_hyperparam(&self, key: &str) -> Option<&f64> {
192 self.hyperparams.get(key)
193 }
194
195 /// Set initial parameter
196 pub fn set_init_param(&mut self, p: Vec<f64>) -> &mut Self {
197 if let Some(x) = self.option.get_mut(&InitParam) {
198 *x = true
199 }
200
201 self.param = p.to_ad_vec();
202 self
203 }
204
205 /// Set maximum iteration
206 pub fn set_max_iter(&mut self, n: usize) -> &mut Self {
207 if let Some(x) = self.option.get_mut(&MaxIter) {
208 *x = true
209 }
210
211 self.max_iter = n;
212 self
213 }
214
215 /// Set optimization method
216 pub fn set_method(&mut self, method: OptMethod) -> &mut Self {
217 self.method = method;
218 self
219 }
220
221 /// Set learning rate for `GradientDescent`
222 pub fn set_lr(&mut self, lr: f64) -> &mut Self {
223 self.hyperparams.insert("lr".to_string(), lr);
224 self
225 }
226
227 /// Set initial lambda for `LevenbergMarquardt`
228 pub fn set_lambda_init(&mut self, lambda_init: f64) -> &mut Self {
229 self.hyperparams
230 .insert("lambda_init".to_string(), lambda_init);
231 self
232 }
233
234 /// Set maximum lambda for `LevenbergMarquardt`
235 pub fn set_lambda_max(&mut self, lambda_max: f64) -> &mut Self {
236 self.hyperparams
237 .insert("lambda_max".to_string(), lambda_max);
238 self
239 }
240
241 /// Main function for optimization
242 pub fn optimize(&mut self) -> Vec<f64> {
243 // Receive initial data
244 let (x_vec, y_vec) = (self.domain.clone(), self.observed.clone());
245 let (p_init, max_iter) = (self.param.clone(), self.max_iter);
246 let safe_f = |p: &Vec<AD>| (self.func)(&x_vec, p.clone()).unwrap();
247 let unsafe_f = |p: Vec<AD>| (self.func)(&x_vec, p);
248
249 // Take various form of initial data
250 let p_init_vec = p_init.to_f64_vec();
251 let y = y_vec.to_col();
252
253 // Declare mutable values
254 let mut p: Matrix = p_init_vec.clone().into();
255 let mut j = jacobian(safe_f, &p_init_vec);
256 let mut y_hat: Matrix = safe_f(&p_init).to_f64_vec().into();
257 let mut jtj = &j.t() * &j;
258 let mut valid_p = p.clone();
259 let mut err_stack = 0usize;
260
261 match self.method {
262 GradientDescent => {
263 let alpha = *self.hyperparams.get("lr").unwrap_or(&1e-3);
264 for i in 0..max_iter {
265 let h = alpha * j.t() * (&y - &y_hat);
266 let p_cand = &p + &h;
267 match unsafe_f(p_cand.data.to_ad_vec()) {
268 Some(value) => {
269 p = p_cand;
270 valid_p = p.clone();
271 err_stack = 0;
272 j = jacobian(safe_f, &p.data);
273 y_hat = value.to_f64_vec().into();
274 }
275 None => {
276 if i < max_iter - 1 && err_stack < 3 {
277 p = p_cand;
278 err_stack += 1;
279 } else {
280 p = valid_p;
281 break;
282 }
283 }
284 }
285 }
286 }
287
288 GaussNewton => unimplemented!(),
289
290 LevenbergMarquardt => {
291 let mut chi2 = ((&y - &y_hat).t() * (&y - &y_hat))[(0, 0)];
292 let mut nu = 2f64;
293 let lambda_0 = *self.hyperparams.get("lambda_init").unwrap_or(&1e-3);
294 let lambda_max = *self
295 .hyperparams
296 .get("lambda_max")
297 .unwrap_or(&f64::MAX.sqrt());
298
299 let mut lambda = lambda_0 * max(jtj.diag());
300
301 for i in 0..max_iter {
302 if lambda > lambda_max {
303 println!(
304 "Caution: At {}-th iter, lambda exceeds max value: {}",
305 i + 1,
306 lambda
307 );
308 break;
309 }
310
311 let h: Matrix;
312
313 let b_lu = (jtj.clone() + lambda * jtj.to_diag()).lu();
314 if b_lu.det() == 0f64 {
315 break;
316 }
317 let b = b_lu.inv();
318 h = b * j.t() * (&y - &y_hat);
319
320 let p_temp = &p + &h;
321 match unsafe_f(p_temp.data.to_ad_vec()) {
322 Some(value) => {
323 let j_temp = jacobian(safe_f, &p.data);
324 let y_hat_temp: Matrix = value.to_f64_vec().into();
325 let chi2_temp = ((&y - &y_hat_temp).t() * (&y - &y_hat_temp))[(0, 0)];
326 let rho = (chi2 - chi2_temp)
327 / (h.t()
328 * (lambda * jtj.to_diag() * h.clone() + j.t() * (&y - &y_hat)))
329 [(0, 0)];
330 if rho > 0f64 {
331 p = p_temp;
332 valid_p = p.clone();
333 err_stack = 0;
334 j = j_temp;
335 jtj = &j.t() * &j;
336 y_hat = y_hat_temp;
337 chi2 = chi2_temp;
338 lambda *=
339 max(vec![1f64 / 3f64, 1f64 - (2f64 * rho - 1f64).powi(3)]);
340 nu = 2f64;
341 } else {
342 lambda *= nu;
343 nu *= 2f64;
344 }
345 }
346 None => {
347 if i < max_iter - 1 && err_stack < 3 {
348 p = p_temp;
349 err_stack += 1;
350 } else {
351 p = valid_p;
352 break;
353 }
354 }
355 }
356 }
357 }
358 }
359 let error_temp = &y - &y_hat;
360 self.error = ((error_temp.t() * error_temp)[(0, 0)] / y.row as f64).sqrt();
361 p.data
362 }
363}